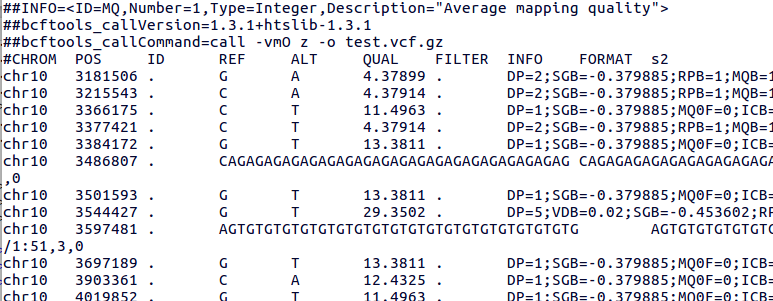
Subsetting with `bcftools filter file1.vcf.gz -r chr#` fails for majority of chromosomes · Issue #774 · samtools/bcftools · GitHub
Possible to annotate the FILTER from an annotation VCF file, as an INFO/TAG in the target? · Issue #1187 · samtools/bcftools · GitHub

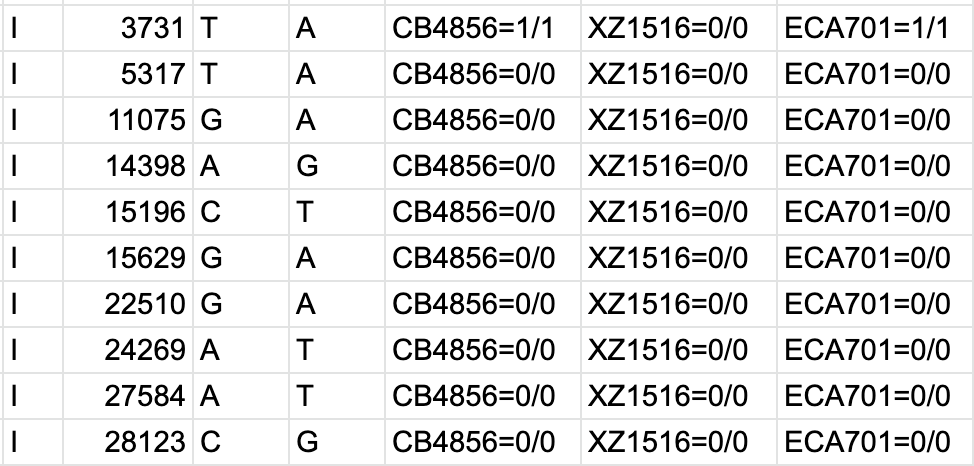
bcftools filter -e 'GT="het"' recognize GT 2/2 as heterozygous · Issue #1268 · samtools/bcftools · GitHub
Comparing -min-DP in vcftools with filter -i 'FORMAT/DP>10' in bcftools · Issue #1384 · samtools/bcftools · GitHub

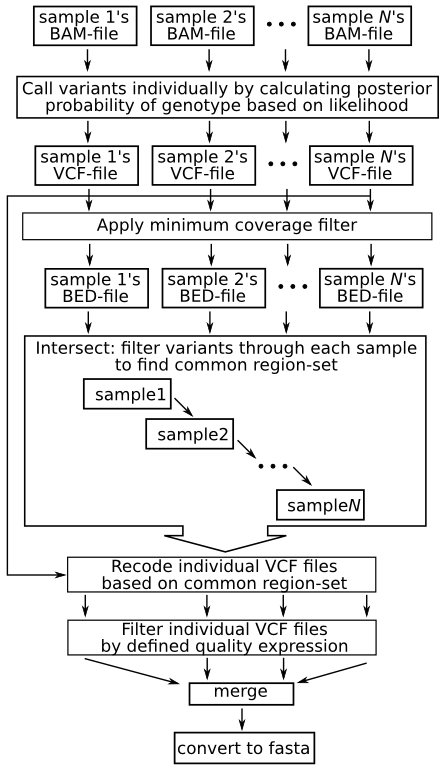
SNP calling and filtering pipelines used in our experiments. (A) In... | Download Scientific Diagram
bcftools filter -e 'GT="het"' recognize GT 2/2 as heterozygous · Issue #1268 · samtools/bcftools · GitHub

Protocol for unbiased, consolidated variant calling from whole exome sequencing data - ScienceDirect
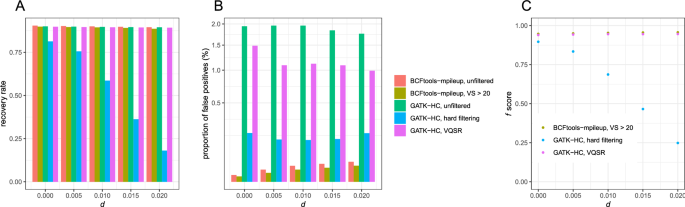
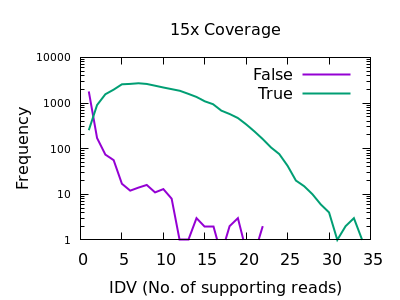
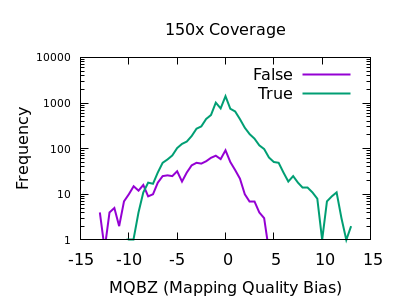
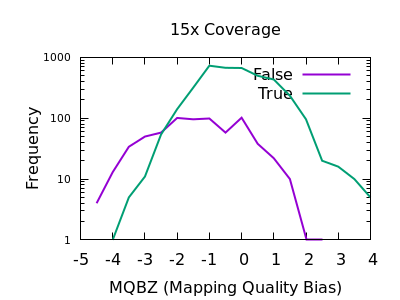
The evaluation of Bcftools mpileup and GATK HaplotypeCaller for variant calling in non-human species | Scientific Reports

bcftools view | bcftools tutorial on how to count the number of snps and indels in a vcf file - YouTube

bcftools view | bcftools tutorial on how to count the number of snps and indels in a vcf file - YouTube